Antimicrobial resistance in small animals: Is there a problem? (Proceedings)
Increasingly, the veterinary practitioner will find that the favorite drug of choice is no longer a viable option.
Increasingly, the veterinary practitioner will find that the favorite drug of choice is no longer a viable option. The role of resistance in therapeutic failure of antimicrobials is well established. However, what is less clear is the impact antimicrobial resistance has in the empirical selection of antimicrobial drugs in the small animal patient, whether the targeted illness be critical or not. This reflects, in part, the limited information available to the veterinarian for the assessment of resistance. This, in turn, reflects the predominant dependence of the small animal clinician on historical data as a guide to antimicrobial therapy. Yet, historical data supporting infections and their cause is often anecdotal in nature, and seldom discriminates infecting microbes from commensal organisms. Empirical therapy will continue to be the predominant basis for antimicrobial selection until more cost-effective, (e.g. molecular-based) diagnostic tests are made available for rapid in-hospital identification of infecting microbes and a description of their susceptibility to drugs. Antimicrobials are among the most common and most important prescribed drugs for the critical care patient and selection generally is made empirically. Yet, the advent of antimicrobial resistance has altered the approach to their use. The goal of antimicrobial therapy includes not only the safe eradication of infection, but now must minimize the advent of resistance. The critical care patients (CCP) is particularly at risk for the development of infections and thus resistance. The risk of nosocomial infection is 5 to 10 fold higher in the critical care ward compared to the hospital-at-large population.. The risk of infection increases due to bacterial translocation, the use of invasive procedures, and the presence of foreign surfaces conducive to bacterial colonization (eg, catheters). Patients are often immunocompromised. Cardiovascular, renal and hepatic dysfunction or responses alter all aspects of drug disposition, increasing the risk of either adverse drug events or therapeutic failure. Polypharmacy increases the risk of adverse drug events and drug interactions. Finally, the sense of urgency accompanying therapeutic decision making generally leads to empirical antimicrobial use.
Antimicrobial resistance might be inherent to the microorganisms, or acquired, either through chromosomal mutations (vertical) or transfer of genetic information (horizontal or shared). Shared mechanisms of resistance include transduction, transformation and conjugation. Development of antimicrobial resistance is facilitated by several factors including duration of hospital stay and patient health. However, among the most important is exposure to antibiotics. The normal flora of the gastrointestinal tract is extremely diverse, with anaerobes predominating. Among the aerobes, E coli are the major Gram negative and Enterococcus the major Gram positive organisms. Environmental microbes maintain an ecological niche through suppression of the competition by section of antibiotics. As such, commensal organisms are constantly being exposed to antibiotics. However, the microbe producing the antibiotic, as well as surrounding normal flora, are resistant to the antibiotic. Thus, genes for resistance develop along with genes directing antibiotic production and organisms are "primed" to develop resistance. The use of synthetic antimicrobials is less likely to induce resistance, perhaps because there is less risk of previous exposure of the flora to these drugs. Rapid microbial turnover in the gastrointestinal tract supports the development of resistance by assuring active DNA and thus mutation potential (see previous discussion). Chromosomal (DNA) mutations (10-14 to 10 -10 per cell division) are DNA mistakes that have been missed by bacterial repair mechanisms. These mistakes occur spontaneously and randomly, whether or not the antibiotic is present. Every time an antibiotic is administered, the normal flora is exposed to varying concentrations of the drug. If the mutation that confers resistance occurs in the presence of the antibiotic, the surviving mutant can progress to a single step mutation, conferring a low level of resistance (see mutant prevention concentration). The MIC of the organism is likely to increase. Further microbial turnover can lead to multi-step mutations and rapid emergence of high-level resistance (eg, unlimited regulation of beta lactamase production) characterized by increasingly higher MIC. Stepwise mutations can lead to specific resistance (ie, to a single drug or drug class), exemplified by clinical resistance to fluorinated quinolones which reflects stepwise mutation in DNA gyrase. However, information for non-specific mechanisms that is shared among organisms can result in multiple organisms developing multiple drug resistance. Microflora of the GI tract can serve as reservoir of resistance genes; a single drug, via integrons, plasmids and transposons facilitate the rapid transfer of multiple drug resistance among organisms.
Shared resistance among bacteria reflects the ability of bacteria to incorporate extrachromosomal DNA carrying the information for resistance from other (including non-self) organisms. Extrachromosomal DNA (including plasmids and bacteriophages) encode for resistance to multiple drugs and can be transmitted vertically (to progeny) or horizontally, across species and genera. Transposons are individual or clusters of resistance genes or repeated DNA sequences (two conserved DNA segments on either side of a resistant gene. Transposons move resistance genes back and forth between chromosomes to plasmids. Integrons are able to capture genes encoding antimicrobial resistance and, unlike transposons, code for an integrase gene. The genes for resistance are located on a gene cassette that can exist as free, circular DNA, but unlike plasmids, can not be replicated or transcribed. Ultimately, the integron must re-integrate into bacterial DNA. As such, bacterial resistance is extremely mobile, and can spread rapidly. Transposons and integrons appear to have played a major role in the advent of multidrug (extreme) resistance isolates of Enterococcus faecalis, Pseudomonas aeruginosa, and Staphylococcus aureus in humans. Among the mechanisms by which genetic resistance information is shared is (sexual) conjugation. Conjugation occurs particularly in Gram negative organisms, and often is accompanied by genetic material that confers bacterial pathogenicity as well as altered metabolic functions. Enterococcus and selected other Gram positive bacteria transfer resistance to glycopeptides through conjugative transposons. Transduction, which requires a specific receptor, involves transfer of information by a bacterial virus (bacteriophage) and is implemented especially Staphylococcus. Resistance, including methicillin, can be transferred between coagulase negative and positive Staphylococcus . Transformation involves transfer of naked DNA from one lysed bacterium; this mechanism of transfer tends to be limited to (in humans) pneumococci meningococci.
The ability of organisms to develop resistance to an antimicrobial varies with the species and strain. Many organisms remain predictably susceptible to selected drugs (eg, Brucella, Chlamydia); whereas others are becoming problematic (Pasteurella multocida). Others have proven be a therapeutic challenge because of resistance that rapidly impairs efficacy of even new antimicrobials (E coli, Klebsiella pneumoniae, Salmonella, Staphylococcus aureus, Streptococcus pneumonia). In general, these organisms have developed multidrug resistance (MDR). Emergence of extended spectrum extended spectrum beta-lactamases (ESBL) is an example of the relentless adaptive nature of microbes toward designer drugs intended to preclude the advent of resistance. Currently, over 340 distinct β-lactamase enzymes are produced by Gram-negative, gram-positive, and anaerobic organisms.. The most prevalent β-lactamases are Class A penicillinases and cephalsporinases, including clinically-relevant TEM-1 or SHV-1 enzymes found in E. coli and Klebsiella peumoniae, and PC1 produced by Staphylococcus aureus. TEM-1 and SHV-1 confer high level resistance to penicillins but generally not cephalosporins or carbapenems. Cefotaxime and ceftazidime initially were assumed indestructible by β-lactamases. However, their high-level use has been associated with the induction and selection for in multiple-resistant coliforms, particularly in those organisms that produce TEM and SHV. The ESBLs are encoded by large plasmids that can confer the information between strains as well as different species of organisms. The gene mutation confers resistance to newer cephalosporins including cefotaxime, ceftazidime and ceftriaxone, as well as cefpodoxime, or 4th generation including cefepime (no longer marketed in the USA); cefipime has been cited as possibly being effective against ESBL.. Monobactams (ie, aztreonam) also are targeted by ESBL. The effect on cephamycins (eg, cefoxitin, cefotetan) is less clear. The impact on clavulanic acid and sulbactam is not clear, although their use in place of cephalosporins appears to reduce the emergence of ESBL and may reduce the emergence of other resistant pathogens such as Clostridium difficile and vancomycin-resistant enterococci.. The ESBL are most commonly found in Klebsiella spp, E. coli or Proteusmirabilis (3.1-9.5%), but they also have been detected in other members of the Enterobacteriaceae and in Pseudomonas aeruginosa. Decreases cephalosporin usage reduces the advent of ESBLs. Because the presence of ESBL may depend on the size of the inoculum at the site of infection, detection requires special testing procedures. The presence of ESBL should be suspected with organisms resistant to cefotaxime but susceptible to β- lactam/β- lactamase combinations. The detection of an isolate with ESBL in a patient with a serious Gram-negative bacillary infection should lead to the use of a carbapenem. However, a novel carbapenemase also has been described has been isolated in Serratia, Klebsiella pneumoniae and E. cloacae.

Feline and canine isolates
Multidrug resistance is now considered the normal response to antibiotics for Gram positive cocci pneumococci, enterococcus and staphylococci. Among these, Staphylococcus is considered most problematic: it is intrinsically virulent, is able to adapt to many different environmental conditions, and it tends to be associated with life threatening infections. In humans, mortality associated with S. aureus bacteremia is 20 to 40%; it is now the leading cause of nosocomial infections in human medicine. Unfortunately, methicillin resistant Staphylococcus (MRSA) is now a community acquired infection in humans, and its recent acquisition of virulence factors has increased the risk of morbidity and mortality. The term MRSA refers to those Staphylococci organisms that are resistant to semi-synthetic beta-lactams, including methicillin. The term was coined in the early 60's when these penicillinase resistant drugs were relatively new. Staphylococcus resistance to penicillin appeared as early as 1942; by the late 1960's, more than 80% of medically relevant isolates were resistant to penicillin. Today, more than 90% of isolates (human) produce penicillinase. Although low level resistance can be overcome by administration of a "protector" such as clavulanic acid, high level resistance also involves production of a penicillin binding protein that has a low affinity for the beta-lactam ring, which can not be overcome by protection. The gene conferring methicillin resistance has been detected in methicillin-resistant Staphylococcus organisms (including intermedius) infecting dogs. Antibiotics are associated with induction, selection and propagation of MRSA. The wide use of cephalosporins, in particular, may have contributed significantly to the advent of MRSA; indeed, increase in MRSA in human patients receiving cephalosporins indicates that it is no longer simply a hospital acquired infection. Detection of MRSA on C&S generally is based on resistance to oxacillin, which is more stable in disks used for testing. Over the 30 to 40 years since the methicillin resistant classification was coined, the infections associated with the organisms have lead to increasing mortality and morbidity. Susceptibility to a number of alternative antimicrobials, including fluorinated quinolones, aminoglycosides, and glycopeptides (vancomycin) is now decreasing. E. coli is another organisms which appears to develop resistance rapidly when expose to selected antibiotics. As with MRSA, the advent of resistance by E. coli as well as other gram-negative organisms has been associated with increased cephalosporin use. Resistance often leads to MDR. More disconcerting, resistance developed by E coli appears to be easily conferred to and from more pathogenic organisms, such as Salmonella. E coli is among the organisms that has developed resistance to the fluorinated quinolones. Even a single dose of a fluorinated quinolone has been associated with changes in the resistant pattern of commensal coliforms in human patients.. Fluorinated quinolone resistant E coli has been documented in the urinary tracts and other tissues of dogs. Previous work by the author found approximately 30% of E coli (n=300, the majority cultured from the urinary tract) resistant to all veterinary fluorinated quinolones and ciprofloxacin. Interestingly, the MIC distribution of isolates was bimodal: isolates were either quite susceptible (MIC50 < 0.06 μg/ml) or very resistant: the MIC 90 was > 64 (g/ml (compared to a breakpoint MIC of 4 μg/ml), suggesting high level multistep resistance. Further, resistance in E. coli cultured from the urine of dogs admitted to a veterinary teaching hospital was characterized by over 40% resistance to drugs most commonly selected empirically, including amoxicillin-clavulanic acid, cephalexin, potentiated sulfonamides and fluoroquinolones. Indeed, the only drugs to which isolates were predictably susceptible included nitrofurantoin and aminoglycosides. Although isolates were susceptible to selected 3rd generation cephalosporins, isolates had not been tested for the presence of ESBLs and as such, the susceptibility should be suspect. A prospective study was performed during the summer of 2005. A follow-up study has confirmed a high incidence of resistance to selected drugs in selected geographical regions. Canine and feline pathogenic E. coli isolates (n=350) were collected from veterinary diagnostic laboratories (9; 4 commercial and 5 Veterinary Teaching Hospitals) in different geographical areas of the country. Isolates were obtained from the laboratories by veterinary practitioners; thus each isolate was collected from a dog or cat presumed to be spontaneously infected. Each isolate was subjected to susceptibility testing by the principal investigator using the Epsilon Test (E-test™) (following guidelines promulgated by the Clinical Laboratory Standards Institute; CLSI) to determine susceptibility versus resistance and the minimum inhibitory concentration [MIC]. Drugs for which susceptibility was determined included amoxicillin (A), amoxicillin-clavulanic acid (X), doxycycline (D), enrofloxacin (E), cefpodoxime (P), trimethoprim-sulfadimethoxine (T), and gentamicin (G). Overall resistance to any one drug ranged from 11% (west) to 47% (south). Overall antimicrobial susceptibility for all isolates, regardless of region, in order of increasing susceptibility was A (49.4%), X (58.3%), P (74.6%), D (75.1%), E (76.4%), T (78.7%), and GN (84.2%) (Figure). For quantitative data, the MIC90 (μg/ml) the MIC90 for isolates generally exceeded the CLSA breakpoint MIC or each drug. Interestingly, whereas 32 isolates were resistant to only amoxicillin and amoxicillin/clavulanic acid, only 2 isolates were resistant only to enrofloxacin, suggesting that resistance to beta-lactams is generally single drug (or drug class), versus resistance to enrofloxacin generally is associated with multidrug resistance.
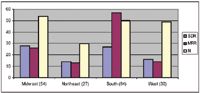
Midwest, Northeast, South, West
Nosocomial organisms are opportunists, probably most commonly from the environment, that cause infections as a result of medical treatment, usually in a hospital or clinic setting. Nosocomial infections are formally defined as infections arising after 48 hr of hospital admission. Because bacterial colonization by nosocomial isolates hospital occurs in the upper respiratory tract, gastrointestinal tract, urogenital tract, and skin of many patients within a few days of hospitalization, nosocomial infections not surprisingly most commonly occur in the respiratory or urinary tract or skin. Infection frequently is associated with invasive procedures. Isolates causing nosocomial infections generally are In some human intensive care units, selected isolates are characterized by resistance prevalence of 86%; resistance results in increased morbidity, mortality, and increased costs. Because nosocomial organisms are characterized by complex resistant patterns, their effective therapy often requires more expensive and potentially toxic drugs and selection clearly should be based on C&S . Nosocomial infections in veterinary critical care patients has been reviewed. The organisms associated with noscomial infections in dogs and cats have been many and diverse, varying with the report. Included (but not exclusively) are Serratia spp, Staphylococcus spp, Streptococcus spp, Klebsiella spp, Enterococcus spp, and E. coli. As in humans, predisposing factors have included catheters and previous antimicrobial therapy.
Percent susceptible
Actions should be taken to avoid antimicrobial resistance, not only for the patient but also for the medical community. Because previous antimicrobial therapy is one of the most important factors associated with resistance, approaches which minimize indiscriminant antimicrobial use will be important. Avoidance of antimicrobial resistance is an overwhelming consideration and will require a different paradigm than has driven antimicrobial use in the past. Probably the single most important first step in judicious antimicrobial use and avoiding resistance is questioning/confirming the need for therapy. This is no small task, being fraught with the lack of effective diagnostic aids. Probably the most common– and least correct mindset is that our directive of "above all else do no harm" is being followed with the use of antimicrobials in the absence of infection. It is paramount that actions be taken that stay the hand. Once the decision to use the antimicrobial is made, efforts should focus on assuring that concentrations adequate to kill the infecting microbe are achieved at the site of infection. Thus, the second step in antimicrobial is identifying the (susceptible) target such that the spectrum of the chosen drug can be as narrow as possible. Historically, empirical therapy has been based on the most likely organisms to infect the site of infection, and the subsequent (presumed) response of the microorganisms to first choice antimicrobials. However, the historical data upon which these choices are base, quite frankly, is lacking in scientific scrutiny. More to the point, as has been addressed, microbes will not sit idly by when faced with exposure to antimicrobials and will develop mechanisms of resistance. Few studies have focused on the success of "guessing" the correct infecting organism in a critical patient; previous work has suggested that clinicians are incorrect up to 50% of the time. In our hospital, that statistic probably would reflect selection of ampicillin as the beta-lactam of choice. Hospitals would be prudent to generate an antibiogram each year to facilitate identification of the bugs most commonly infecting patients and not only the incidence of resistance versus susceptibility, but the level of susceptibility (ie, MIC). Note that once an antibiotic is begun, should the choice be incorrect, C&S data collected before the drug was begun may no longer be relevant to the patient. The third action that can be taken to reduce the incidence of resistance is to ensure that adequate drug concentrations are reached at the site of infection. Adequate implies bacterial killing, not simply inhibition. DEAD BUGS DON'T MUTATE! Attention to PDC is important not only for efficacy, but also in order to reduce the risk of resistance. Generally, a microbial populations with a density is ≥ 107 colony forming units (CFU) will generate sufficient spontaneous mutations that an organism may become more resistant toward any antibiotic being used to treat a patient. Routine culture and susceptibility data, which is based on 105 organisms, is not likely to detect these first step mutants that have undergone the first step of resistance. If antimicrobial therapy is designed to achieve the MIC of the cultured organism the growth of all isolates except the first step mutants will be inhibited. Subsequent growth of the mutant survivors will yield a population of organisms that are characterized by reduced susceptibility although the organism is not yet identified as resistant. When this new population grows to ≥ 107 CFU, a second step mutation will occur, yielding CFU now characterized by a high level of resistance. The MIC of these second step mutants are not likely to be reached at recommended doses. The mutant prevention concentrations (MPC) is defined as the highest MIC identified in a population (≥ 107 ) of susceptible organisms, thus including those organisms that have undergone the first step mutation. Simplistically, whereas the MIC indicates the concentration of drug which inhibits the growth of the organism, the MPC indicates the concentration of drug that minimizes the advent of its resistance. The MPC, rather than the MIC, should be the targeted concentration of drug at the site of infection in order to overcome first step of mutational resistance. Interestingly, achieving drug concentrations below the MIC of the infecting organism is not likely to facilitate the advent of resistance; because concentrations are too low to inhibit the growth of most organisms, selection pressure will be avoided. Clinical evidence supports the development of multistep resistance in human patients receiving antimicrobial therapy designed to achieve the MIC of the infecting organism. A first step mutation has been demonstrated in bacterial prostatitis in men receiving only three days of ciprofloxacin at doses designed to achieve the MIC but not the MPC. Prophylactic use of ciprofloxacin for urologic procedures has been associated with the advent of resistant E. coli. In the author's (DMB) experience, dogs afflicted with urinary tract infections, particularly if treated with lower doses of FQ, subsequently develop infections associated with the same organism, but characterized by an MIC two or more tube dilutions higher than the original infecting organism. A similar scenario has been described in humans. In humans, the design of dosing regimens are now focusing on avoidance of the second step mutation and high level resistance by targeting the MPC rather than the MIC of the infecting organism. This approach underscores the importance of collecting culture and susceptibility data in animals previously treated with antimicrobials. E coli, Staphylococcus aureus, and Pseudomonas aeruginosa are examples of organisms for which concentrations high enough to target single step mutants should be achieved. Unfortunately, determining the MPC of an isolate cultured from a patient requires culture techniques based on ≥ 107 organisms, which currently is not possible.. In one study, the ratio of MPC to MIC for various FQs toward human pathogens ranged from a low of 6 to 10 for E. coli (ATCC 8739) but 23 to 50 and as high as 125 for selected drugs toward Staphylococcus aureus (ATCC 6538). Increasingly in human antimicrobial dosing regimens are focusing on a shorter duration of therapy with higher doses. Exceptions to shorter duration might be made for organisms characterized by slow growth. The mutant selection window can be narrowed if more than two bacterial sites are targeted, such as might occur with combination antimicrobial therapy, or with drugs that simultaneously target more than one site, such as the fluoroquinolones. However, organisms often exhibit many different levels of susceptibility to fluoroquinolones; use of drugs at concentrations near the lower boundary of the window nurtures the development of resistance because so many mutants exist in the normal population.
The most significant mechanism by which bacterial resistance is likely to be reduced is modification of antimicrobial prescribing behaviors. However, also important will be implementation of behaviors that are designed to reduce patient risk such as length of hospital stay, and design, implementation of and adherence to infection control practices. Intensive care units have implemented a number of techniques intended to reduce antimicrobial resistance. Hospital strategies involve a multi-tiered approach, including: 1. improving infection control (eg, selective decontamination procedures, prevention of horizontal transmission via handwashing, glove and gown use, alternatives to soap, and improving the workload and facilities for health care workers); 2. improving appropriate antimicrobial use (eg, including less ideal strategies such as strict adherence to prescribed formularies, setting limits on the duration of antimicrobial therapy; potentially reasonable strategies such as requiring prior approval for use of certain antibiotics such that proper use can be verified; and more rationale strategies such as narrowing the spectrum of empiric antibiotics, and rotating the use of antimicrobial drugs on a regular schedule); 3. primary prevention by decreasing length of hospital stay, decreased use of invasive devices, and newer approaches such as selective digestive decontamination and vaccine development.4. Improved information systems technology also plays a role as hospitals increasing are communicate with one another, approaching the issue as a global rather than hospital concern.
Each proposed or implemented strategy has theoretical benefits and limitations, but data on their efficacy in controlling antimicrobial resistance are limited. Among the proposed strategies, antimicrobial cycling or rotation appears to be receiving the most interest. This strategy involves the regularly scheduled substitution of a class of antibiotics (eg, aminoglycosides with extended spectrum penicillins), or within a class, an different member (ie, amikacin versus gentamicin) that exhibits comparable antimicrobial activity.. The latter tends to be more difficult and potentially less effective because cross resistance within a class is not unusual. The basis for this intervention is the recognition that increased use of any drug is accompanied by an increase in selection pressure and thus increase in the incidence of resistance; and the belief that decreased use will be associated with a return to the cycle with efficacy intact. The time that must elapse between cycles (removal and reintroduction of the drug) varies with the amount of resistance exhibited toward the drug prior to its removal. The duration of drug use should be sufficiently short to assure that a clone of organisms does not develop resistant to the drug. As appealing as this strategy is, the timing of cycles is likely to be hospital dependent. However, evidence does support that resistance will decline toward a drug even if the drug is substituted with another member of the same class (eg, gentamicin vs amikacin; ceftazidime vs piperacillin/tazobactam). The specifics (proper cycling length, classes of drugs for which this approach is effective, impact of diseases, etc) have yet to be determined. Even though the strategies may not significantly decrease outcome measurements such as duration of treatment or costs or length of hospital stay for human medical patients, they appear to have at least slowed the progression of negative changes. It is likely that strategies that will prove most effective are those reflecting a combination of approaches.
Among the more rational paradigms for antimicrobial control is antibacterial de-escalation, an approach to empirical antibacterial use in the hospital setting for patients with serious bacterial infections. Antibacterial de-escalation attempts to balance the need to provide appropriate, initial antibacterial treatment while limiting the emergence of antibacterial resistance. The goal of de-escalation is to prescribe an initial antibacterial regimen that will cover the most likely bacterial pathogens associated with infection while minimizing the risk of emerging antibacterial resistance. The three pronged approach includes narrowing the antibacterial regimen through culture, assessing the susceptibility for dose determination and choosing the shortest course of therapy clinically acceptable. That such methods are making headway has been demonstrated in isolated studies. For example, judicious antimicrobial used combined with restricted use of ceftazidime led to decreased antimicrobial resistance to beta-lactams in generally in a human teaching hospital environment; a 44% reduction in use was associated with a 12 to 24% reduction in resistance to ceftazidime and 18 to 32% reduction in piperacillin and 12 to 20% reduction in imipenem resistance.. Restricted fluoroquinolone use appears to reduce the prevalence of fluoroquinolone resistance in E. coli but appears to have less impact of resistance prevalence in S. aureus or Pseudomonas aeruginosa.
Podcast CE: A Surgeon’s Perspective on Current Trends for the Management of Osteoarthritis, Part 1
May 17th 2024David L. Dycus, DVM, MS, CCRP, DACVS joins Adam Christman, DVM, MBA, to discuss a proactive approach to the diagnosis of osteoarthritis and the best tools for general practice.
Listen