Antimicrobial therapy and urinary tract infections: part 1-the other bad E. coli (Proceedings)
The advent of antimicrobial resistance is increasingly limiting therapeutic options in human and veterinary medicine. The ability of organisms to develop resistance to an antimicrobial varies with the species and strain.
"It is unwise to underestimate an adversary that has had a three billion year evolutionary head start" (Sayers, 2004). The advent of antimicrobial resistance is increasingly limiting therapeutic options in human and veterinary medicine. The ability of organisms to develop resistance to an antimicrobial varies with the species and strain. Among the most adaptable organisms is E. coli. Discovered in 1885 by a the ediatrician Theodore Escherich, it was originally discovered in neonatal fecal samples. Dr. Erich recognized E. coli was acquired at birth and remained with us till death, with strains coming and going. It is the most thorourghly understood microbe, and is critically important as a research tool. Through E. coli, we have come to understand such diverse activities as intermediary metabolism, DNA replication and RNA transcription, protein synthesis and genetic recombination. Indeed, recombinant products would not be possible without E. coli. Escherichia coli, a member of the family Enterobacteriaceae, is a lactose fermenter, causing a distinct color on diagnostic agar. It is the predominant facultative anaerobe (in the normal intestine of both humans and many warm-blooded animals), playing a major role as normal microflora.1-2 However, it also is ubiquitous in the environment, as is recognized by its appearance as contaminants in food stuffs. It has or acquires genes that encode for flagella, making it mobile. Its presence in the environment is used as a sentinel of environmental contamination. Referred to as the "cockroach" of microbes because of its adaptability, E. coli rapidly divides, potentially doubling its population every 20 minutes. Further, it is highly mutagenic, with spontaneous mutations occuring in of 1 per 100 thousand to 1 per billion new progeny (assume 1 gm of feces contains 100 million E coli) thus assuring opportunity for spontaneous mutation even in the absence of stimuli, such as drugs. E coli occurs in many hosts, and is present in multiple tissues, thus also being associated with diseases: the gastrointestinal tract, brain (humans: meningioencephalitis) and the urinary tract are major sites of pathology. Diarrhea, UTI, and sepsis are example sequelae. However, it is not appropriate to consider E. coli only as a pathogen. Indeed, E. coli enjoys a bit of a "Dr. Jeckell, Mr. Hyde" designation. A common research tool, K12 is a "wild type" isolate that expresses neither virulence or resistance and often serves as a control. In contrast, the most notorious E coli is O157:H7 (defined by antigens and genetic sequences that define it as a distinct strain); this isolate has been associated with contaminated food and is a cause of a hemolytic-uremic syndrome. The pathogenicity of these E. coli reflect its ability to acquire virulence factors.

Virulence and resistance are characteristics of concern for the patient infected with this organism. Virulence refers to the ability of an organism to cause disease whereas resistance refers to the ability of the organism to avoid harm, although clinically we tend to use the term in reference to avoidance of harm caused by antimicrobials. Resistance and virulence are often perceived to emerge simultaneously. Recently, however, studies suggest that resistance may be associated with a reduced potential for invasiveness and a decrease in the presence or the expression of some virulence factors in E. coli. Virulence implies the ability of an organism to cause harm to a patient and should not be confused with resistance; indeed, the two may be mutually exclusive. Although enteric E.coli are generally nonpathogenic, the presence of virulence factors in some strains allows classification of E. coli into 6 groups capable of causing enteric disease: Extraintestinal E coli (ExPEC) is a more recently classified pathotype; UPEC (uropathogenic) are included in this category. Although much attention has focused on the impact of enteropathogenic E. coli (e.g., OH157:H7) in human medicine, other E. coli, particularly ExPEC, also are associated with morbidity and mortality. These include sepsis associated (SEPEC) and neonatal meningitis associated (NEMEC) E. coli. The ExPEC appear to easily colonize the gastrointestinal tract, potentially displacing commensals, and eventually emerging as infectious organisms in other body tissues, particularly in the urinary tract. These factors are of concern not only to the small animal clinician, but increasingly are contributing to a potential public health concern. UPEC are ExPEC that acquire virulence factors necessary for survival of the organism outside of the gastrointestinal tract. In humans, E. coli is associated with 90% of UTI in otherwise healthy persons. It is also responsible for approximately 50% of nosocomial UTI. Emerging statistics indicate the same is true in dogs. The pathophysiology of human and canine UTI associated with E. coli is quite similar. Isolates causing UTI are from the gastrointestinal tract. Ascending infection up the urethra to the bladder may be further complicated by infection in the ureter and kidney if the isolate contains P fimbrae or the K antigen. Infection may also occur in the prostate. Once in the bladder, several virulence factors facilitate survival in the bladder. Initial release of cytotoxic materials destroy uroepithelial cells, facilitating penetration, but also providing nutrients for the microbes. Virulence factors facilitate scavenging of iron, a necessity in the bladder. However, among the host defenses is its own scavenging of iron, preventing the microbe from accessing the mineral. However, as a testament to the adaptability of E. coli is its ability to generate another virulence factor that inhibits the hosts ability to scavenge iron. Critical to successful infection is the ability of E. coli to adhere to the uroepithelium, thus protecting the microbe from bulk urine flow, among the most important defenses of the bladder. Adhesins involve mannose receptors which are recognized by the E. coli ; however, isolates with the K antigen do not detect mannose receptors. Once the E. coli has adhered to the uroepithelium, the production of biofilm will protect it from damage by host cells and antimicrobials. Biofilm communities are complex and sophisticated. Biofilm will potentially protect microbes such that they become senescent, and thus less susceptible to antimicrobial therapy. Such cells may remain under the uroepithelial cells until they are exfoliated, only to become active again such that infection continues. The genes that code for virulence are located on pathogenicity islands; generally,such organisms do not carry genes for resistance.
Studies have demonstrated transfer of resistant E. coli between animals and humans, a fact which was documented as early as 1975. Canine E. coli strains have been demonstrated to be phylogenetically similar to pathogenic strains infecting humans. Over 15% of environmental canine fecal deposits contain E. coli strains related to virulent human strains. Evidence that pets and owners share E. coli is increasing as has been demonstrated in studies within family members, including pets. However, what is not clear is if zoonoses or reverse zoonoses predominated, although it is more likely that owners are infecting or sharing isolates than the opposite. Nontheless, public health concerns mandate that antimicrobials be used judiciously; further, when considering options for treating or preventing UTI, attention must be given not only to the antimicrobial history of the veterinary patient, but also the household members-be they 4 legged or 2 legged.
Resistance.
The gastrointestinal environment is conducive to development of resistance. Environmental microbes maintain an ecological niche by suppressing competition through secretion of antibiotics. As such, commensal organisms are constantly being exposed to antibiotics. However, the microbe producing the antibiotic, as well as surrounding normal flora, are resistant to the antibiotic. Thus, genes for resistance develop along with genes directing antibiotic production and organisms are "primed" to develop resistance. Microflora of the GI tract can serve as reservoir of resistance genes. Exposure to antimicrobials may facilitate survival of isolates that have either spontaneously mutated or acquired resistant through other means. Resistance may be easily conferred to other potentially more virulent organisms. E. coli rapidly develops resistance, particularly that associated with multiple drug resistance (MDR) when exposed to selected antimicrobials. More disconcerting, resistance is easily conferred to more pathogenic organisms. In human medicine, E. coli has developed resistance to the fluorinated quinolones, beta-lactams, or both: it is among the Gram negative organisms that secrete extended spectrum beta-lactamases (ESBL). Emergence of extended spectrum extended spectrum beta-lactamases (ESBL) is an example of the relentless adaptive nature of microbes toward designer drugs intended to preclude the advent of resistance. The ESBLs are encoded by large plasmids that can confer the information between strains as well as different species of organisms. The gene mutation confers resistance to newer cephalosporins includeing cefotaxime, ceftazidime and ceftriaxone, as well as cefpodoxime, or 4th generation including cefepime (no longer marketed in the USA); cefipime has been cited as possibly being effective against ESBL.The impact on clavulanic acid and sulbactam is not clear, although their use in place of cephalosporins appears to reduce the emergence of ESBL and may reduce the emergence of other resistant pathogens such as Clostridium difficile and vancomycin-resistant enterococci. The ESBL are most commonly found in Klebsiella spp, E. coli or Proteus mirabilis (3.1-9.5%), but they also have been detected in other members of the Enterobacteriaceae and in Pseudomonas aeruginosa. Resistance to fluoroquinolones has also been well characterized. Normally associated with point mutations in topoisomerases (DNA Gyrase and Topoisomerase IV), such resistance is, like beta-lactmases, within class. However, in the presence of continued drug, efflux pumps appear to be induced. Such pumps serve to remove toxic compounds from the organism, including antimicrobials. At least 5 efflux pump systems have been characterized; they are associated with porins. They are characterized by broad substrate specificity, thus can impart multidrug resistance.The the culture (above) and the antibiogram below typify the patterns of resistance that can emerge in animals which have received fluoroquinolones and in which resistance has emerged.
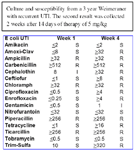
E coli resistance in isolates associated with urinary tract infections is particularly well described . One study demonstrated the gastrointestinal emergence of quinolone -resistant E. coli genetically distinct from infection-causing strains in patients treated with ciprofloxacin. Like virulence factors, transfer of resistance genes in isolates between animals in humans may present a public health risk, as was recognized by the FDA by 1999. E. coli develops resistance both vertically and horizontally. Shared resistance reflects the ability of bacteria to incorporate extrachromosomal DNA carrying the information for resistance from other (including non-self) organisms. Extrachromosomal DNA (including plasmids and bacteriophages) encode for resistance to m ultiple drugs and can be transmitted vertically (to progeny) or horizontally, across species and genera. In general, resistance carried by plasmids "comes and goes", meaning the presence of the drug may increase the likelihood of the plasmid being present, in large copy numbers; removal of the drug may be associated with resolution of the resistance.
We have demonstrated the impact of E. coli MDR through several of our studies. In healthy normal dogs, we have demonstrated the impact of 10 mg/kg amoxicillin bid and 5 mg/kg enrofloxacin once daily orally for 7 days on fecal E. coli. For either drug, 100% of E coli became resistant to the drug, expressing high level (more than 8 times the MIC breakpoint). For amoxicillin, this resistance was limited to beta lactams and occasionally tetracyclines or sulfonamides; resistance tended to resolve by the 3 week study end period. Enrofloxacin resistance howeer, not only was multidrug resistance, but tended to persist. The relationship between enrofloxacin resistance and multidrug resistance was also demonstrate din a pilot surveillance study of approximately 400 E. coli pathogens collected form dogs or cats. The pattern of resistance varied regionally, being as much as 50% to amoxicillin or amoxicillin clavulanic acid and in the south, approximately 30% to enrofloxacin. Although the number of isolates resistant to beta lactams only (expressing single drug resistance) was high, single drug resistance to enrofloxacin was rare. If resistance was expressed to enrofloxacin, it was multidrug in nature. We have an ongoing study involving 3000 isolates throughout the United States sponsored by the Morris Animal Foundation and Idexx laboratories. Currently, regional differences in resistance continue to persist.Overall resistance is greatest to cephalexin.

The Three "DE"s
Regardless of the organism, the most significant mechanism by which bacterial resistance is likely to be reduced is implementation of behaviors that are designed to reduce patient risk such as length of hospital stay, and design, implementation of and adherence to infection control practices. Consider the three DE-s:
Descalate (1)
Because previous antimicrobial therapy is one of the most important factors associated with resistance, approaches which minimize indiscriminant antimicrobial use will be important. Examples of human strategies include improving appropriate antimicrobial use (eg, including less ideal strategies such as strict adherence to prescribed formularies, setting limits on the duration of antimicrobial therapy); potentially reasonable strategies such as requiring prior approval for use of certain antibiotics such that proper use can be verified; and more rationale strategies such as narrowing the spectrum of empiric antibiotics, and rotating the use of antimicrobial drugs on a regular schedule); primary prevention by decreasing length of hospital stay, decreased use of invasive devices, and newer approaches such as selective digestive decontamination and vaccine development. Probably the single most important first step in judicious antimicrobial use and avoiding resistance is questioning/confirming the need for therapy. This is no small task, being frought with the lack of effective diagnostic aids. Probably the most common– and least correct mindset is failure to recognize that we are in conflict with our directive of "above all else do no harm" if we use antimicrobials in the absence of infection. De-escalation includes taking actions that stay the hand in reaching for drugs if they are not really necessary. For urinary tract infections, increasingly the need for treating asymptomatic bacteria is questioned. What constitutes an infection may not depend only on the inoculum size (e.g, 1000 CFU/ml) but the presence of clinical signs.
Design(2)
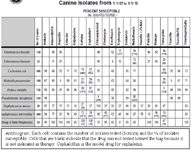
Dosing regimens should be designed to assure that adequate drug concentrations are reached at the site of infection to kill, not simply inhibit, microbial growth. DEAD BUGS DON'T MUTATE! Once the decision to use the antimicrobial is made, efforts should focus selecting a drug to which the bug is most susceptible. A practice-based antibiogram (see above) may be helpful. If an animal has not been exposed to antimicrobials, the chances are improved that any infecting pathogens are among the susceptible isolates. A narrow spectrum is preferred to a broad spectrum drug. Once the drug is chosen, design focuses on the dosing regimen to assure that concentrations adequate to kill the infecting microbe are achieved at the site of infection. This involves not only selecting the drug to which the isolate is most susceptible (and the drug most likely to reach the target site), but also designing the dosing regimen based on the MIC, potentially the MPC and time or concentration dependency of the drug (see parts II and III if relevant). An antibiogram (see figure; top number is % susceptible, bottom number of each cell is number tested) generated for each practice can support empirical selection of antimicrobial drugs although increasingly, culture is indicated in all but the antimicrobial naïve patient (this includes direct - or indirect through a household member - exposure). (Note that squares without information are drugs which should not be tested toward that bug).
Detergent (3)
Hospital strategies include: improving infection control (eg, selective decontamination procedures, prevention of horizontal transmission via handwashing, use of disinfectants, glove and gown use, alternatives to soap, and improving the workload and facilities for health care workers), and identification of specific areas for treatment of potentially infectious agents (ie, bandaging areas that can be easily cleaned). Increasingly "detergent" should be applied to the patient and its home. For example, recurrent infections might be reduced if successful initial therapy is coupled with cleansing of the environment in which the pet is located such that it is not continued to be exposed to the infecting bug. For UTI infections, this may become particularly important in that urine contaminates the environment.
Podcast CE: A Surgeon’s Perspective on Current Trends for the Management of Osteoarthritis, Part 1
May 17th 2024David L. Dycus, DVM, MS, CCRP, DACVS joins Adam Christman, DVM, MBA, to discuss a proactive approach to the diagnosis of osteoarthritis and the best tools for general practice.
Listen