Interpreting diagnostic test results (Proceedings)
The innate sensitivity and specificity of a diagnostic test affects its validity, i.e. how well a test reflects the true disease status of an animal.
The innate sensitivity and specificity of a diagnostic test affects its validity, i.e. how well a test reflects the true disease status of an animal. An assay is considered 'validated' if it consistently produces test results that identify animals as being positive or negative for the presence of a substance (e.g. antibody, antigen, organism) in the sample (e.g. serum) being tested and, by inference, accurately predicts the infection status of the tested animals with a predetermined degree of statistical certainty (confidence). Inferences based on these test results can be made about the infection status of the animals. The process of validating an assay is the responsibility of researchers and diagnosticians. The initial development and optimization of an assay, by a researcher, may require further characterization of the performance of the assay by laboratory diagnosticians before field use. Keep in mind that the specific criteria required for assay validation of an infectious disease are elusive and that the process leading to a validated assay is not standardized. Factors that influence the capacity of the test result to accurately infer the infection status of the host are diagnostic sensitivity (Sn), diagnostic specificity (Sp), and prevalence of the disease in the population targeted by the assay.
Diagnostic Sn and diagnostic Sp of a test are calculated relative to test results obtained from reference animal populations of known infection/exposure status to a specific disease agent. Diagnostic Sn describes the proportion of animals with disease that have a positive test result. Diagnostic Sp describes the proportion of animals free of a disease that have a negative test. The degree to which the reference animals represent all of the host and environmental variables in the population targeted by the assay has a major impact on the accuracy of test result interpretation (accuracy is discussed in greater detail below) and applicability of this test in this population.
The capacity of a positive or negative test result to accurately predict the infection status of the animal is a key objective of assay validation. This capacity is not only dependent upon a highly precise and accurate assay and carefully derived estimates of Sn and Sp, but is also strongly influenced by the prevalence of the infection in the targeted population. Without a current estimate of the disease prevalence in that population, the interpretation of a positive or negative test result will be compromised.
When diagnostic tests are dichotomized into 'positive' or 'negative', a cut-offpoint is required. Cut-off points have been determined in several ways. Visual inspection of the frequency distributions of the test results for infected and uninfected animals has been used. The frequency distributions for infected and uninfected animals typically indicate an overlapping region of assay results (the perfect test with no overlap, yielding 100% diagnostic Sn and 100% diagnostic Sp, rarely—if ever—exists). The cut-off is placed at the intersection of the two distributions. This method has the advantage of being simple and flexible, and requires no statistical calculations or assumptions about the normality of the two distributions. Another way to determine a cut-off point is to arbitrarily place it two or three standard deviations greater than the mean of the test values of the unaffected individuals. This approach fails to consider the frequency of disease and the distribution of test results in diseased individuals, and ignores the impact of false-positive and false-negative errors. Another approach is one that identifies 95% of individuals with disease as being test-positive. This ignores the distribution of test results in unaffected individuals, the prevalence of the disease, and all consequences except those that are due to false-negative error. Alternatively, the value that minimizes the total number or total cost of misdiagnoses can be selected. The optimum cut-off point also depends on the frequency distribution of the test variable in the healthy and diseased population, which may be complicated. A cut-off point can also be established by a modified receiver-operator characteristics (ROC) analysis.
When a cut-off point is identified, there is then clearly an inverse relationship between Sn and Sp in a particular test. Individuals for whom the assays value is to the right of the cut-off point are classified as diseased (assuming that the diseased animals have a higher test results than the non-diseased animals), typically a combination of truly diseased animals (true positive; TP) and non-diseased (healthy) animals mis-classified as diseased (false positive; FP). This is the Sn established for the test. With a sensitive test, the proportion (prevalence) of truly diseased animals will be higher in the group tested positive than in the initial population of animals to be tested. Individuals for whom the assay's value is to the left of the cut-off point are classified as healthy, typically a combination of truly non-diseased (healthy) animals (true negatives; TN) and diseased animals mis-classified as healthy (false negative; FN), again the post-test proportion (prevalence) of non-diseased animal will be higher than the proportion in the pretest population. This is the Sp established for the test. If fewer false positives are required, the cut-off point is moved to the right. Diagnostic Sp of the test increases and diagnostic Sn decreases. However, if fewer false negatives are required, the cut-off point is moved to the left. Diagnostic Sn increases and diagnostic Sp decreases.
Calculation of sensitivity and specificity requires an independent, valid criterion—also termed a 'gold standard'—by which to define an animal's true disease status. The diseased and healthy animals to which the 'gold standard' is applied should be representative of the population in which the test is to be applied. Thus, a test is conducted in the general population, and so the 'gold standard' should be applied to a sample of diseased and healthy animals drawn from this population. In contrast, a clinical diagnostic test is run on animals for which there is usually already evidence of disease where the prevalence of the disease is higher, and the test needs to distinguish between animals with the relevant condition (the 'diseased' animals) and those animals with other diseases in your differential disease list (the 'healthy' animals). It follows that the choice of a specific test with a given sensitivity and specificity may be different when it is applied as a test than when it is used as a diagnostic test in a veterinary clinic due to the higher disease prevalence rate in clinically ill animals.
The following example is presented to illustrate the diagnostic sensitivity (Sn) and diagnostic specificity (Sp) of a test in a general population of animals and how they are influenced by the choice of the cut-off point. In this example, a 60 head beef herd has a known 25% prevalence of disease "X" (45 healthy animals; 15 diseased animals).
= 15 Diseased
= 45 Healthy
Test "Y" is conducted looking for disease agent "X" in a sample of blood drawn from each animal in the herd. The disease status of each animal is known. Each animal is classified as diseased or healthy based on the following cut-off point of test "Y" as established by the laboratory conducting the test at left.

Example 1
Let's assume that the laboratory wants to improve the sensitivity of the test in order to identify all diseased animals, as would be the case when attempting to eradicate a disease. In order to accomplish this goal, the test cut-off point must be shifted to the left. The example at right shows that the test cut-off point has been moved to the left sufficiently to result in a test that has 100% sensitivity.

Example 2
Please note that the effort to improve the sensitivity of test "Y" has eliminated the number of false negatives but the trade-off to this action is that the number of false positives increases, i.e. the number of healthy animals misclassified as diseased increases.
Let's consider a different scenario. Assume that the laboratory wants to improve the specificity of the test in order to identify all healthy animals. In order to accomplish this goal, the test cut-off point must be shifted to the right. The example at left shows that the test cut-off point has been moved to the right sufficiently to result in a test that has 100% specificity.

Example 3
Please note that the effort to improve the specificity of test "Y" has eliminated the number of false positives but the trade-off to this action is that the number of false negatives increases, i.e. the number of diseased animals misclassified as uninfected increases.
Accuracy of a Diagnostic Test
The accuracy of a diagnostic test is measured by the proportion of animals correctly identified or classified as healthy (true negative; TN) or diseased (true positive; TP) by the test, i.e. accuracy = (TN + TP) / N, where N is the number of animals sampled. For example, the accuracy of test "Y" at the original test cut-off point was (42 + 14) / 60 = 93.33%.
Predictive Value of a Diagnostic Test
Remember, diagnostic sensitivity (Sn) and diagnostic specificity (Sp) are innate characteristics of a test and (for a defined cut-off point) do not vary. As a veterinarian, you do not have control over the diagnostic Sn and diagnostic Sp of a test itself unless, of course, you were the person that developed the test or you convince laboratory personnel to adjust the cut-off point to make it more sensitive or specific for your situation. Interestingly, employing multiple diagnostic tests is another way to improve overall diagnostic Sn or diagnostic Sp, depending on which one is your focus (to be discussed later). Consequently, the Sn and Sp of a test does not help you decide what a test result means when the disease status of an animal is unknown. The positive and negative predictive value of a test helps understand and interpret test results from a herd with unknown disease status.
Positive Predictive Value or PPV (also referred to as predictive value positive or PVP) estimates the probability that the animal is diseased, given that a test is positive. Negative Predictive Value or NPV (also referred to as predictive value negative or PVN) estimates the probability that the animal is not diseased (healthy), given that a test is negative.
The graph at right illustrates the relationship between PPV and NPV of a diagnostic test, with a given diagnostic Sn (95%) and diagnostic Sp (95%), as the prevalence of disease within the herd changes.
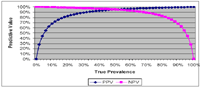
PPV vs. NPV
You should note that PPV and NPV have an inverse relationship as prevalence of disease changes. When the prevalence of disease within a herd decreases, the positive predictive value (PPV) of the diagnostic test with a defined Sn and Sp also decreases. In practical terms, what this means is that as prevalence of disease decreases, out of the 'positive' animals identified by the test, an increasingly larger number of them will be false positives (FP) and fewer true positives (TP). In contrast, the negative predictive value (NPV) of an initial test increases as the prevalence of disease within a herd decreases. In practical terms, what this means is that as prevalence of disease decreases, out of the 'negative' animals identified by the test, an increasingly larger number of them will be truly negative (TN) and fewer and fewer will be false negatives (FN).
Just the opposite occurs as the prevalence of disease within a herd increases; PPV increases and NPV decreases. In practical terms, what this means is that as prevalence of disease increases, out of the 'positive' animals identified by the test, an increasingly larger number of them will be truly positive and out of the 'negative' animals, an increasingly larger number of them will be false negatives.
Estimating True Prevalence of Disease from Diagnostic Test Results
When testing a herd or flock of unknown disease status, the proportion of 'positive' animals out of all animals tested is a measure of test prevalence (also known as apparent prevalence, Papparent) of disease, but not the true prevalence of disease within a herd or flock. However, if diagnostic sensitivity (Sn) and diagnostic specificity (Sp) of the test are known, the true prevalence of disease, Ptrue, can be calculated using Papparent according to the following formula:
Ptrue = (Papparent + Sp – 1) / (Sn + Sp – 1)
For example, if 40% of a herd of 100 animals is 'positive' for disease "X" when tested using a test with .90 Sn and .95 Sp, the true prevalence of disease is estimated to be:
Ptrue = (.4 + .95 – 1) / (.9 + .95 – 1) = .35 / .85 = .4117 or 41.17%
In this example, the calculation (41.7%) for true prevalence of disease actually represents a mean. The true prevalence of disease in this herd lies within a range of values. A confidence interval (CI) can be calculated, which provides a way of expressing the range over which a value is likely to occur. Although any range can be used, the 95% CI is most commonly used in the veterinary literature. A 95% CI means that you are 95% confident that over many samplings, the mean prevalence of disease "X" in this herd will lie within such and such an interval. Alternatively, it means that if you performed 100 samplings of the same number of animals, 95 of the herd's true prevalence values are predicted to lie in this interval and 5 will not. Why? Because the herd prevalence is an estimate of the population's mean it has a 95% probability of being within this interval.
In order to calculate a 95% CI, the mean, variance, and standard deviation of disease prevalence (also referred to as standard error of the proportion) must be available. In the example above, given the true prevalence (equivalent to the mean) of disease of 41.7%, the variance of disease prevalence equals [p(1-p)/n], where p is the proportion of diseased individuals and n is the sample size. Thus, the variance is (.417 X .583)/100 or 0.00243. The standard error of the proportion equals the square root of the variance: v0.00243 = 0.049.
The 95% CI for disease "X" = Ptrue ± 1.96 (standard error of the proportion)
CI = .417 ± 1.96 X 0.049
= .417 ± .096
= .321 to .513
Instead of 41.7%, now 95% confident that true prevalence of disease "X" is between 32.1% and 51.3%).
Screening versus Confirmatory Diagnostic Test
In a typical surveillance scenario dealing with an outbreak of disease, a 'screening test' refers to the testing of a wide cross-section of a population of apparently healthy individuals in order to detect infection (subclinical disease). The test used is usually not aimed at establishing a definitive diagnosis. Rather, the aim is to separate individuals that probably have a disease from those that probably do not. Thus, it is very important for the screening test to be of high diagnostic Sn. The diagnostic Sp of the screening test can be compromised for the sake of achieving high diagnostic Sn. Alternatively, the PPV of a screening test can be improved by sampling only high risk populations, i.e. targeted surveillance of populations likely to have a high rate of infection or disease. Remember, PPV and prevalence of disease have a positive relationship—as prevalence of disease increases so does the PPV of a test and as prevalence of disease decreases so does PPV of the test.
The aim of a 'confirmatory test' is to confirm results derived from other test methods, such as a screening test. Since the reason for using a confirmatory test is most often to confirm that an individual is diseased, a confirmatory test is typically chosen that has high diagnostic Sp—reduces the number of false positives—since the objective is to confirm a positive result, i.e. confirm presence of disease in an individual. In general, a confirmatory test is typically more difficult to perform and takes longer to perform than a screening test, is less readily available within a laboratory system because of additional expertise needed to perform the test, and is more expensive than more commonly used screening tests. A confirmatory test could actually be a 'gold standard'.
However, if the objective is to establish freedom from disease in the individuals being tested, a confirmatory test would be run instead on all negative animals. This test should be one that possesses high diagnostic sensitivity (Sn)—reduces the number of false negatives.
Employing Multiple Diagnostic Tests
The objective of using multiple tests is to improve predictive value. This is the likely way in which a disease outbreak is handled in the field. One approach is to use a series of one or more confirmatory tests after a screening test has been used to initially categorize individuals as positive ('series positive' testing regime) or negative ('series negative' testing regime). Ideally, confirmative tests should be a different biological type than the screening test. In the case of individuals classified as positive, the innate specificity of the confirmatory test should be greater, in an effort to decrease the probability of a false positive occurring. In a 'series positive' testing regime an animal is deemed to be infected with the disease agent in question if it is 'positive' to all tests (Table 1). Otherwise, the animal is deemed to be uninfected. 'Serial positive' testing is essentially asking the animal to prove that it is affected by the disease.

Table 1. Examples of probable disease status in different animals based on 'series positive' testing.
The purpose of 'series negative' testing is to rule out a disease. The innate sensitivity of the confirmatory test should be greater, if at all possible, in an effort to decrease the probability of a false negative occurring. Decreasing the probability of a false negative occurring will decrease the likelihood that a diseased animal will be missed. An animal is deemed to be uninfected with the disease agent in question if it is 'negative' to all tests (Table 2). Otherwise, the animal is considered to be diseased if at least one of the tests is positive. 'Series negative' testing is essentially asking the animal to prove that it is healthy.

Table 2. Examples of probable disease status in different animals based on 'series negative' testing.
In summary, 'series positive' testing:
- Used to rule in a disease
- Improves the overall diagnostic specificity (Sp) and positive predictive value (PPV) of the combined tests—few false positives and more true negatives are identified; greatest predictive value is a positive test result
- Decreases the overall diagnostic sensitivity (Sn) and negative predictive value (NPV) of the test regime—more false negatives and fewer true positives are identified
- Increases the risk of missing a diseased animal
- Most useful if rapid assessment is not necessary, i.e. when time is not critical such as with test and removal programs.
- Also useful to employ when there is an important penalty for false positive results.
In summary, 'series negative' testing:
- Used to rule out a disease
- Improves the overall diagnostic sensitivity (Sn) and negative predictive value (NPV) of the combined tests—few false negatives and more true positives are identified; greatest predictive value is a negative test result
- Decreases the overall diagnostic specificity (Sp) and positive predictive value (PPV) of the test regime—more false positives and fewer true negatives are identified
- Decreases likelihood of missing a disease
- Most useful when a rapid assessment of disease status of individual (diseased) animals is needed or in emergency situations
- Also useful to employ in situations when there is an important penalty for missing a disease (i.e. false negative results).
References
Jacobsen RH. Validation of serological assays for diagnosis of infectious diseases. Rev Sci Tech Off Int Epiz 1998;17(2):469-486.
Thrusfield M. Diagnostic testing. In: Thrusfield M, ed. Veterinary Epidemiology. 2nd ed. London: Blackwell Science Ltd. 1995;266-285.
Smith RD. Statistical significance. In: Smith RD, ed. Veterinary Clinical Epidemiology. 3rd ed. Boca Raton, FL: CRC Press. 2006;137-161.
Martin SW, Meek AH, Willeberg P. In: Martin SW, Meek AH, Willeberg P, eds. Veterinary Epidemiology—Principles and Methods. Ames, IA: Iowa State University Press. 1987;48-76.
Podcast CE: A Surgeon’s Perspective on Current Trends for the Management of Osteoarthritis, Part 1
May 17th 2024David L. Dycus, DVM, MS, CCRP, DACVS joins Adam Christman, DVM, MBA, to discuss a proactive approach to the diagnosis of osteoarthritis and the best tools for general practice.
Listen











